The sleeping Sigma-Factor
A previously unknown mechanism enables bacterial antibiotic resistance
Within their environment, bacteria often encounter various kinds of changes. In order to survive under stress or changing conditions, bacteria have to respond fast and adequate to achieve a physiological adaptive response. This mostly happens by specifically adapting gene expression. Thus, transcriptional regulation is one of the biggest means by which bacteria adapt to external stress conditions.
Initiation of transcription in bacteria only occurs upon the binding of a key component known as the sigma factor (σ factor) to the RNA polymerase (RNAP) core enzyme in order to form the complete and catalytically active holoenzyme. This holoenzyme then recognizes key promotor elements and subsequently enables transcription. During external stress conditions, the primary σ factor is replaced by an alternative σ factor, which differs from the former with respect to the promoter sequences that it recognizes. Thus, holoenzyme formation with this alternative σ factor results in the transcription of corresponding stress-response genes. Among the various classes of alternative σ factors, the most abundant ones are the extracytoplasmic function (ECF) σ factors. Sigma factors are commonly known to be intrinsically active, which means that the bacterial cell has to keep them in an inactive state until their action is warranted.
Novel mechanism of transcriptional regulation
There are several mechanisms for regulating σ factor activity. Usually, alternative σ factors stay retained in an inactive state by sequestration into a complex with an anti-σ factor. Upon a specific stimulus, the inhibitory effect of the anti-σ factor is alleviated and the σ factor is released for interaction with the RNA polymerase.
However, in collaboration with researchers from SYNMIKRO in Marburg, the Max Planck researchers have revealed a previously unknown mechanism of transcriptional regulation, which instead relies on intrinsically inactive σ factors that are unable to bind the RNAP core enzyme. Only upon phosphorylation on a specific residue is the σ factor activated and able to bind the RNAP in formation of the holoenzyme and consequently drive expression of specific genes. An extensive bioinformatics analysis indicated that this transcriptional regulation by σ-phosphorylation is a general mechanism in bacteria, presenting a new paradigm in transcriptional regulation.
“A special property of this mechanism is its modularity”, Research Group Leader Simon Ringgaard explains. “Our discoveries reveal how nature has merged two distinct regulation mechanisms – threonine kinase signaling and regulation of σ factor activity – as joint forces, in order to achieve the ability of environmental adaptation.”
The novel sensing and signaling pathway also regulates antibiotic resistance
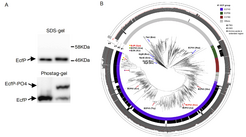
Ringgaard`s model organism itself represents a field of application: Vibrio parahaemolyticus is a severe human pathogen and the leading agent of seafood borne gastroenteritis in the world. The Max Planck researchers identified an ECF σ factor / threonine-kinase pair (named EcfP / PknT) that is responsible for sensing polymyxin antibiotics stress and mediating bacterial resistance towards polymyxin in V. parahaemolyticus. The PknT kinase is activated when cells are treated with polymyxin antibiotics. Activated PknT in turn activates EcfP, which results in expression of genes required for polymyxin antibiotic resistance.
Polymyxins constitute a class of antibiotics that count as a last resort to treat Gram-negative infections. Because antibiotic resistance is a severe public health concern worldwide, it is very important to understand how cells sense and respond to antibiotic treatment. The identification of mechanisms that regulates polymyxin antibiotic resistance in V. parahaemolyticus opens up new avenues of research in fighting this and probably other important human pathogens. Ultimately, the work of the Marburg researchers provides fundamental insights in the regulation of gene expression and cellular adaptation in the entire bacterial kingdom.












