A Killer Microbe as a Living Antibiotic
Predatory bacteria may give rise to novel protein-based anti-microbial substances
Predatory bacteria are native to many microbial communities and have been found in terrestric and aquatic ecosystems, as well as in the human and animal intestine. A research team at the Max-Planck-Institute for Developmental Biology, together with their colleagues from the universities of Nottingham and Bielefeld has now unraveled for the first time the complete genome sequence of a predatory bacterium in order to identify molecular mechanisms that are important when bacteria hunt their own. Insights into this ancient dependency may give rise to novel anti-microbial substances. These substances, however, will not be based on the structures of today’s chemical antibiotics, but rather will be deduced from the protein sequences that become available from the Bdellovibrio genome project. Furthermore, the scientists predict in their publication of this weeks edition of Science magazine that Bdellovibrio may be developed into a therapeutic agent that could be used as a "living antibiotic".
Bdellovibrio bacteriovorus is a fascinating predatory bacterium that attaches specifically to certain other bacteria in order to invade them. Once it has entered its prey, it begins to consume the host cell from the inside. Using Bdellovibrio’s genomic information the life cycle of this unique bacterium can now be studied for the first time on a molecular level.
In the free-living phase of its life cycle Bdellovibrio swims at high speed while locating areas of high prey concentration by use of its chemosensory system. Once it has collided with a prey cell, Bdellovibrio stays reversibly attached to it while verifying its suitability for invasion. In the presented model, the recognition mechanism is likely to involve one of several pilus systems that produce long retractable fibers that allow Bdellovibrio to pull itself into close proximity with its prey. By using a lytic cocktail that is capable of degrading lipids, proteins and carbohydrate molecules, Bdellovibrio then generates an opening in the cell wall of the prey. Via a pulling motion the predator navigates itself in the "periplasmic space" between the outer and inner membrane of the prey cell.
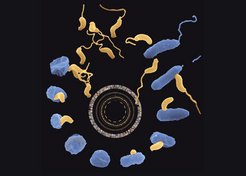
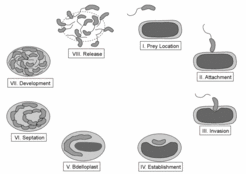
Bdellovibrio can remain encysted at this stage, while the entry pore has been sealed and the prey cell remains viable. Most commonly, however, Bdellovibrio immediately enters its growth phase in which it depends on the prey’s amino acids. The amino acids and other nutrients are made available to the invader by the degradation of biopolymers in the cytoplasm of the prey cell and are subsequently transported into the Bdellovibrio cell. In this way the cytoplasm of the prey is entirely consumed, while the Bdellovibrio cell elongates. Upon exhaustion of all prey resources Bdellovibrio’s life cycle continues, with the bacteria differentiating back into as many as 15 motile cells, which seek out and attack new prey.
The genomic analysis of Bdellovibrio showed that this organism uses a large variety of lytic enzymes, which can degrade complex biopolymers of the prey, such as proteins, carbohydrates, DNA and RNA. The researchers will attempt to identify the targets in the prey cell that have proven to be successful points of attack in this million-year-old prey-predator relationship. The lytic enzymes acting on cellular systems that are not targeted by conventional chemical antibiotics are thereby especially interesting.
Far reaching anti-microbial strategies aim at using Bdellovibrio as a "living antibiotic". This seems feasible, as Bdellovibrio is not capable of infecting eukaryotic cells, in particular mammalian cells. Moreover, it was shown in animal experiments that Bdellovibrio only has a weakly immunogenic surface, which does not produce serious life threatening reactions in test animals. These attributes, together with the facts that certain Bdellovibrio strains show a very narrow prey spectrum and are capable of penetrating the same tissues as may human-pathogens, gives promise to the development of novel anti-microbial strategies.

