A stomach pathogen "Out of Africa"
Max-Planck scientists show that prehistoric human migrations are reflected by the global population structure of Helicobacter pylori.
A tiny bacterium (Helicobacter pylori) that lives in our stomachs can reveal the migration patterns of our ancestors. Scientists in the groups of Mark Achtman at the Max Planck Institute for Infection Biology, Berlin and Sebastian Suerbaum at the University of Würzburg have investigated the gastric bacterial pathogen Helicobacter pylori from 27 geographic and ethnic sources and deduced its modern and ancestral population structure by novel genetic methods. In the latest issue of Science (Science, March 7 2003), they report that geographic isolation of the human hosts allowed the evolution over millennia of distinct ancestral bacterial populations in Africa, the Near East, Central Asia and East Asia. Subsequent prehistoric migrations of various human ethnic groupings resulted in the evolution of seven modern populations that are now merging due to globalization.
The molecular clock rate for mammalian evolution can be calibrated on the basis of the fossil record. For bacteria, which lack a fossil record, the rate of evolution is easiest to estimate when they co-evolve with their hosts. Helicobacter pylori is a bacterium that colonizes the stomach mucosa of half of the global human population. Colonization with H. pylori can last for decades in the absence of treatment and result in gastric and peptic ulcers as well as predisposing for lymphoma. H. pylori is largely transmitted within families and does not spread epidemically. Furthermore, these bacteria are characterized by high genetic diversity, about 50fold that of humans, and DNA sequences differ with the geographical source of isolation.
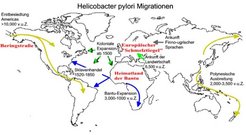
Together with partners from six other American and French institutions, the microbiologists from Berlin and Würzburg have now shown that H. pylori has accompanied human migrations since ancient times. 370 haplotypes were identified by sequencing 1418 polymorphic nucleotides within H. pylori isolated from 27 distinct ethnic and geographic sources. Population genetic analysis of the sequence diversity revealed that the bacteria belong to seven modern populations and subpopulations. A novel population genetic method was then implemented to reconstruct the nucleotides present in five ancestral bacterial populations that evolved in Africa, the Near East, Central Asia and East Asia. By comparison between the ancestral and modern populations, it has been possible to reconstruct how H. pylori has been distributed globally by specific human migrations (Figure).
During colonization of the stomach by multiple strains of H. pylori, free DNA from lysed bacteria is imported by horizontal genetic exchange, resulting in mosaic genomes where patches of nucleotides have been imported. Modern European H. pylori resulted from the genetic fusion by recombination of two ancestral populations that were independently imported from Central Asia and the Near East. Similarly, distinct bacterial subpopulations evolved during the thousands of years of isolation that followed the Polynesian expansion to the Pacific, the Siberian migrations to the Americas and the Bantu expansion in Africa.
In many areas, native bacteria now compete for colonization of the human stomach with foreign bacteria that were imported in the last few hundred years due to recent human migrations, such as the European colonization of the Americas, Africa and Australia and the slave trade with the Americas.
These new results are potentially important for medical treatment of gastric ulcers and cancer. Genetic differences between distinct bacterial populations may correlate with differences in virulence and antigenic structure, which could affect the efficacy of particular drug regimens and vaccine strategies. Furthermore, the dates of individual human migrations that resulted in subdivisions between bacterial populations can now be used to calibrate H. pylori’s molecular clock rate.
In a commentary on these results in the same issue of Science, the english microbiologist Brian G. Spratt discusses the possibility that genetic diversity within H. pylori might help resolve between alternative and controversial interpretations of individual human migrations. Although favoring the use of "direct measures of human ancestry", he points out that the "approaches used by Falush et al. can be extended to humans to study the levels of social interaction required for the acquisition of H. pylori".
