Homeostasis is vital for bacteria
Structural investigation by cryo-electron microscopy on a bacterial potassium ion-proton symporter
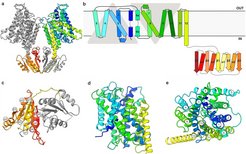
a) KimA dimer viewed from the membrane, with one monomer coloured in grey and the other in a rainbow, from blue (N terminus) to red (C terminus). b) Schematic representation of the topology of KimA. Each KimA monomer is composed of 12 TM helices, organized in a 5 + 5 inverted structural repeat and two additional, C-terminal TM helices located at the periphery of the dimer. The last TM helix extends into the cytoplasm and positions the soluble domain below the TM domain of the second monomer. c) The cytoplasmic domain of a KimA monomer is formed by a parallel, five-stranded β-sheet sandwiched by four α-helices, forming a continuous 10-stranded β-sheet in the dimer. d) Side view and e) top view of the membrane domain of a KimA monomer.
A balanced potassium concentration is essential for the survival of all organisms from bacteria to humans. For bacteria, controlled potassium uptake is often difficult because they are continuously exposed to strong fluctuations of environmental conditions. The potassium ions can only be specifically taken up by specific membrane transport proteins; the cell membrane is impermeable to potassium ions.
With the help of cryo-electron microscopy the structure of KimA was determined up to a resolution of 3.7 Å, the first structure of a KUP (K+ uptake permease) family member. Of particular interest is the region that can bind potassium ions. Here are conserved amino acids within the KUP family, which can bind K+ and H+. This was shown by point mutations.
Three potassium ion binding sites were identified in the transmembrane region of the protein. In the present structure the KimA homodimer has an inwardly closed, trans-inhibited conformation.
The cryo-EM map and the model were deposited in the wwPDB with accession codes EMD-10092 and 6S3K, respectively.
