Molecular time travel
A simple rule of biochemistry drives the evolution of useless complexity
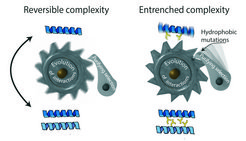
A ratchet-like mechanism makes the evolution of useless complexity irreversible. Left: When proteins first evolve to interact with each other, those interactions often have no functional benefit and can be lost without consequence. Right: the portions of the protein that are now buried in the interface quickly accumulate hydrophobic mutations (yellow sticks) to levels that would be deleterious if they were exposed to the watery environment of the cell. Purifying selection then prevents reversal back to the simpler state.
Most proteins in our cells form specific complexes with other proteins, a process called multimerization. Like other kinds of complexity in biology, multimers are usually thought to persist over evolutionary time because they confer some functional benefit that is favored by natural selection. “How complexity evolves is one of the great questions of evolutionary biology,” said senior author Joseph Thornton, professor of human genetics and ecology and evolution at the University of Chicago. “The classic explanation is that elaborate structures must exist because they confer some functional benefit on the organism, so natural selection drives ever-increasing states of complexity. Clearly in some cases complexity is adaptive, like the evolution of the eye: complex eyes see better than simple ones. But at the molecular level, we found that there are other simple mechanisms that drive the build-up of complexity.”
The research team, led by Thornton and Georg Hochberg, who is now a group leader at the Max Planck Institute in Marburg, Germany, set out to study the evolution of multimerization in a family of proteins called steroid hormone receptors, which assemble into pairs (called dimers). They used a technique called ancestral protein reconstruction, a kind of molecular “time travel,” Thornton said, that allowed them to recreate ancient proteins in the lab and experimentally examine how they were affected by mutations that happened hundreds of millions of years ago.
Dimers with deleterious mutations
To their surprise, they found that the ancient proteins functioned no differently when assembled into a dimer than if they had never evolved to dimerize at all. There was nothing useful or beneficial about forming the complex. The explanation for why the dimeric form of the receptor has persisted for 450 million years turned out to be surprisingly simple. “These proteins gradually became addicted to their interaction, even though there is nothing useful about it,” explained Hochberg. “The parts of the protein that forms the interface where the partners bind each other accumulated mutations which were tolerable after the dimer evolved, but which would have been deleterious in the solo state. This made the protein totally dependent on the dimeric form, and it could no longer go back. Useless complexity became entrenched, essentially forever.”
By analyzing a huge database of protein structures, the researchers showed that the genetic code makes this "evolutionary ratchet" universal. They found the principle to be realized at a fundamental level - the interplay of fat and water solubility. Mutations that make a protein’s surface more oil-soluble impair its folding, so purifying selection removes them if they occur in solo proteins. But once a protein evolves to multimerize, the parts of the surface that form the interface become hidden from water, so oil-soluble mutations are no longer deleterious; they become invisible to selection and can become a fixed part of the protein. The multimer is then entrenched, because returning to the solo state would expose the hydrophobic interface.
Useless complexes
The hydrophobic ratchet, operating on thousands of proteins over hundreds of millions of years, could drive the gradual accumulation of many useless complexes inside cells. “Some complexes surely have important functions, but even those will be entrenched by the hydrophobic ratchet, making them harder to lose than they would otherwise be,” Hochberg said. “With the ratchet constantly operating in the background, our cells have built up a massive stock of entrenched complexes, many of which may never have performed a useful function, or long ago ceased to do so.”
Future directions include investigating whether or not interactions other than dimerization may be the result of entrenchment. “This was a story about proteins dimerizing with other copies of themselves, which is a super common process,” said Thornton. “But there are lots of other interactions in cells, and we think it’s possible that some of those may have accumulated during evolution because of a similar kind of acquired dependence on molecular complexity.”