From restriction enzymes to CRISPR-Cas9
CRISPR-Cas9 is not the first method available to scientists for modifying DNA; it is by far, however, the easiest to use. With CRISPR-Cas9, the crRNA/tracrRNA sequence or an artificial guide RNA indicate where the DNA can be cut. It is relatively easy for scientists to produce different sequence variants from RNA molecules in the laboratory. In the past, this involved the adaptation of entire proteins or parts of proteins – a process that is considerably more complicated than working with small RNA molecules. For this reason, compared with all previously available methods, it is much easier to ‘program’ CRISPR-Cas9 at a particular cutting point on the DNA strand.
Restriction enzymes
Various types of endonucleases – enzymes that can cut DNA – were already known before CRISPR-Cas9. The discovery of restriction enzymes in the early 1970s heralded a new age in molecular biology. These enzymes recognize characteristic DNA sequences and cut them. Bacteria and Archaea can also use these enzymes to locate foreign DNA and render it harmless.
Scientists used the restriction enzymes to, among other things, cut DNA at particular locations and insert new genes at the cutting sites. However, it is difficult to determine in advance exactly where the gene will be inserted, as the recognition sequences of most restriction enzymes are just a few base pairs long and often arise several times in a genome. Moreover, the specificity of a restriction enzyme is dependent on environmental conditions.
For this reason, scientists looked for ways of improving the accuracy of restriction enzymes and of modifying them so that they could recognize a sequence that is unique in the genome. Meganucleases are an example of such unique sequences. The design of such enzymes that can recognize longer DNA sequences is particularly complex, however, as DNA recognition and cutting take place in one protein segment.
Another result of this research are zinc finger nucleases.
Zinc finger nucleases
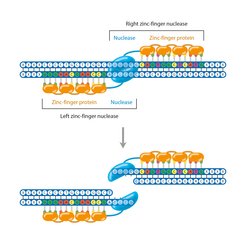
These artificial enzymes consist of a sub-unit that recognizes the desired DNA sequence, and a DNA-cutting part of a restriction enzyme. Several zinc ions are bound to the amino acid chain in the DNA-binding domain. This forms loops as a result (‘zinc fingers’) which can bind to DNA. Each zinc finger recognizes a characteristic sequence of three base pairs. Three aligned zinc fingers are therefore sufficient to accurately recognize a particular location in the genome.
But producing such zinc finger nucleases requires a lot of skill and effort, as does modifying the zinc fingers so that they dock at a desired DNA sequence. For example, it is not sufficient to know the individual recognition sequence of several zinc fingers and combine them to make a cut at the desired site, because they influence their individual recognition sequences mutually. Although there are different methods available for doing this, only a few research laboratories in the world are able to produce customized zinc finger nucleases.
TALENs
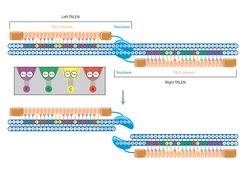
Transcription activator-like effector nucleases (TALENs) are somewhat easier to produce than zinc finger nucleases. They also consist of a DNA-cleaving subunit of a restriction enzyme and a sequence recognition component known as the TAL effector. TAL effector proteins are usually used by bacteria to activate genes in plant and animal cells so that they can infect them more easily.
TAL effectors consist of up to 33 repeats of a sequence that is usually 34 amino acids long. Two of these amino acids are extremely variable and determine the letters of the genetic code – the bases – recognized by the sequence. Due to the high number of repeats, TAL effectors can bind to both longer and unique sequences in the genome.
Thanks to the simple correspondence of the two variable amino acids with the letters of the genetic code, it is easier for scientists to develop TAL effectors for certain DNA sequences than zinc fingers. However, it remains a complex task.

